Next: PDF Estimation using HMMs Up: PDF Estimation using Gaussian Previous: CR Bound analysis Contents
names={'ENGY','TIME'};
min_std = [.1 .1];
NMODE=1;
gparm1=init_gmix(data1,NMODE,names,min_std);
The first line assigns names to the two dimensions. We
have chosen to call them "ENGY" and "TIME".
The next line assigns the gparm1=gmix_trainscript(gparm1,data1,150);The log likelihood (Q) is printed out at each iteration along with the number of modes. Log likelihood would monotonically increase, if not for the pruning, splitting, and merging operations. Use of the CONSTRAINT method of covariance conditioning will also affect the monotonicity. It may be verified, however, that calls to software/gmix_step.m with BIAS=1 will result in monotonic likelihood increase, with the possible exception of numerical errors at the very end of the convergence process. Whenever mode splitting occurs, the message “Adding a mode .." is printed. Whenever mode merging occurs, the message “Merging ..." is printed. Because in this example, we have initialized with just one mode, mode splitting is more likely that merging, although is is possible that after several modes have been split, they can be re-merged. This causes a “fight" between software/gmix_kurt.m which tries to split, and software/gmix_merge.m, which tries to merge. In software/gmix_trainscript.m, it is arranged to allow time between splitting and merging so that the E-M algorithm can settle out. Otherwise, newly merged modes could be quickly split, or newly split modes could be quickly merged.
Once software/gmix_trainscript.m converges, you should should see the graph on the left of Figure 13.13.
These plots are produced by software/gmix_view2.m. This utility is perhaps the most useful visualization tool for high-dimensional PDF estimation. It allows the data scatter plot to be compared with the marginalized PDF on any 2-dimensional plane.
Marginalization
is a simple matter for Gaussian mixtures.
Let
![]() . To visualize
on the
. To visualize
on the ![]() plane, for example, we would
need to compute
plane, for example, we would
need to compute
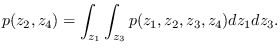
gparm_out = gmix_strip(gparm_in, [2 4]);where the second argument indicates that we want to retain the second and fourth dimensions. Using this method, the marginal distribution of any 2-dimensional plane is easily computed. Stripping is handled automatically by software/gmix_view2.m.
Press once more and the intensity plot is replaced by a contour plot of the modes (on the right of Figure 13.13. The contour plot is obtained by the fourth argument to software/gmix_view2.m. The complete syntax of software/gmix_view2.m is
[p,xp,yp]=gmix_view2(gparm1,data1,idx1,idx2,[do_ellip],[M],iplot);where idx1, idx2 are the indexes of the dimensions requested and do_ellip is an optional argument that, if equal to 1 (default=0), produces a contour plot of each mode instead of an image. M is an optional parameter that defines the number of resolution cells for each dimension of the plot (default=60). The optional parameter iplot can be set to zero if only the outputs p,xp,yp are wanted. These outputs provide the output PDF grid that can be plotted by imagesc(xp,yp,p). In the example, since the data is 2-dimensional to begin with, there is no dimension reduction performed by software/gmix_view2.m.
Information about the Gaussian mixture may be printed by calling software/gmix_show.m. This information, which includes the mode weights, means, and determinants, can be directly compared with Figure 13.13. The true means are (2,3) and (.5,.5), and the true determinants are 1.44 and 1.0, respectively. Generally, if the algorithm results in just 2 modes, the parameters agree very closely. The inclusion of a third or fourth mode makes it difficult to see the correspondence. But, nevertheless, the PDF approximation is good as evidenced by the intensity plot. You can run software/gmix_example.m again and each time the result will be a little different. But always, the intensity plot and the PDF approximation is excellent.
Press the key once more and Figure 13.14 will be plotted.
This figure shows the 1-dimensional marginals for each dimension displayed along with the histograms. It is the result of calling software/gmix_view1.m. The calling syntax is[pdf,xp,h,xh]=gmix_view1(gparm,data,idx,nbins);If called without any output arguments, the plot will automatically be generated. Input idx is an array of indexes for the dimensions desired. For more than one index, multiple plots are produced. Input “nbins" is the histogram size.
Press a key once more and Figure 13.15. This figure
demonstrates the function gmix_cond (See Section
13.2.6) which
creates conditional PDFs from the original Gaussian mixture.
Rather than using Bayes rule explicitly to compute
the conditional PDF of x given that ![]() ,
,
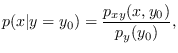
Press a key once more and Figure 13.16 is shown. This figure plots the original data again in green and some synthetic data created by software/gmix_makedata.m in red. This demonstrates a convenient aspect of GM approximation: generating synthetic data is simple.
Press a key once more and Figure 13.17 appears.
We now have two data sets. We will now build a classifier using Gaussian mixtures. The first step is to train a second parameter set on the second data set. This time, we will use the top-down approach by initializing with 15 mixture modes. This time, there will be alot of merging and purging going on, but less splitting! Press a key again and the training starts. When complete, you should see Figure 13.18.To classify, it is necessary to compute the log-likelihood of test data. This is done using software/lqr_evp.m. The name of the subroutine was not thought up logically, but evolved from (l)og-likelihood (ev)aluation from the (p)arameters, and the fact that the (QR) decomposition is involved in the covariance estimates! The calling syntax is
loglik = lqr_evp(gparm1,data1,0);If the third argument was 1, the routine would return a matrix of log-likelihoods where each column is from one of the mixture modes. The zero forces the modes to be combined with the apropriate weights into the GM approximation. The ROC curve is shown in Figure 13.19. Refer to the listing for details.