Conditioning the Covariances
Conditioning the covariances is accomplished without explicitly
computing
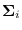 as well. As mentioned in
Table 13.1, step 5, there are two methods, the
BIAS and CONSTRAINT methods. The BIAS method is simpler.
On the other hand, the CONSTRAINT method delivers a better
PDF estimate because the covariances are not biased
and appears to converge faster. But, it may
interfere with the monotonic
increasing property of the E-M algorithm,
i.e. that the total log-likelihood always goes up,
but this is still an unresolved issue.
Both methods are based on the idea of
independent measurement error in the elements of
as well. As mentioned in
Table 13.1, step 5, there are two methods, the
BIAS and CONSTRAINT methods. The BIAS method is simpler.
On the other hand, the CONSTRAINT method delivers a better
PDF estimate because the covariances are not biased
and appears to converge faster. But, it may
interfere with the monotonic
increasing property of the E-M algorithm,
i.e. that the total log-likelihood always goes up,
but this is still an unresolved issue.
Both methods are based on the idea of
independent measurement error in the elements of
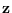 . Let
. Let 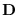 be a diagonal covariance matrix
with
be a diagonal covariance matrix
with
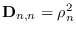 .
The two methods differ in how they regard
.
The two methods differ in how they regard
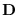 .
The BIAS method assumes
.
The BIAS method assumes 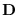 is an
a priori estimate of
is an
a priori estimate of
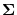 , while the CONSTRAINT method
assumes
, while the CONSTRAINT method
assumes 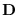 is a measurement error
covariance.
is a measurement error
covariance.
The BIAS method is implemented by adding
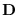 to the newly formed
covariance estimate. But, because we do not work
with
to the newly formed
covariance estimate. But, because we do not work
with
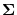 directly, it is necessary
to implement the conditioning as follows: Let
directly, it is necessary
to implement the conditioning as follows: Let
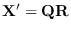 .
The upper triangular matrix
.
The upper triangular matrix 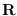 is retained
and
is retained
and 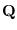 is discarded. Next,
we form the matrix as shown below for the case
is discarded. Next,
we form the matrix as shown below for the case 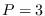 :
:
It may be verified that
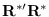 is the same as
is the same as
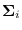 with the diagonal
adjustments. Next, the QR-decomposition of
with the diagonal
adjustments. Next, the QR-decomposition of 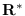 is computed
and the upper triangular part is stored.
is computed
and the upper triangular part is stored.
The CONSTRAINT method assumes that
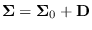 where
where
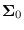 is an arbitrary covariance.
Let the eigendecomposition
of
is an arbitrary covariance.
Let the eigendecomposition
of
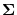 be
be
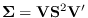 .
Clearly, then
.
Clearly, then
Thus, the diagonal elements of 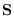 can be no smaller than the
square root of the diagonal elements of
can be no smaller than the
square root of the diagonal elements of
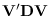 .
Note that
.
Note that 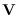 and
and 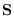 may be obtained
from the SVD of the Cholesky factor of
may be obtained
from the SVD of the Cholesky factor of
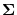 :
:
and
It is implemented in this way in
software/gmix_step.m(tmpvar corresponds to 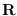 ):
):
[U,S,V]=svd(tmpvar,0);
S = diag(S);
S = max(S,sqrt(diag( V' * diag(minvar) * V )));
tmpvar = U * diag(S) * V';
[q,tmpvar] = qr(tmpvar,0);
where the last two steps re-construct 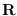 ,
then force it to be upper triangular.
,
then force it to be upper triangular.
Consider the following example. Data was created using
a mixture of 2 Gaussians using the code segment below:
%
% produce data that is from two Gaussian populations
%
fprintf('Creating data : ');
N=4096;
mean1=[2 3]';
cov1= [2 -1.6; -1.6 2];
mean2=[1.3 1.3]';
cov2= [.005 0; 0 .005];
x1 = chol(cov1)' * randn(DIM,N);
x1=x1+repmat(mean1,1,N);
x2 = chol(cov2)' * randn(DIM,N);
x2=x2+repmat(mean2,1,N);
data1 = [x1 x2];
Next, a GM parameter set was initialized with 2 modes
with random starting means. Next,
software/gmix_step.mwas iterated 50 times using the BIAS and the CONSTRAINT
method. This experiment was repeated 9 times.
In each trial, the same starting point was used
for both methods. The results are plotted in Figures
13.2 and 13.3.
Figure 13.2:
Convergence performance of the BIAS and
CONSTRAINT methods. The CONSTRAINT method is consistently
faster and achieves a higher log-likelihood every time.
|
|
Figure:
Typical results of training using
the BIAS (left) and CONSTRAINT (right) methods. Each method used
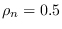 . Note that for the BIAS method, the
covariance of the large mode is too fat, but for the
CONSTRAINT method it is correct.
For the small mode, the mode size is much
smaller than
. Note that for the BIAS method, the
covariance of the large mode is too fat, but for the
CONSTRAINT method it is correct.
For the small mode, the mode size is much
smaller than 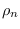 and therefore
both methods produce similar results, as would be expected.
and therefore
both methods produce similar results, as would be expected.
|
|
Note that the BIAS method has
covariances that are biased and appear
somewhat larger than necessary.
In every case, the CONSTRAINT method converged
faster and achieved a higher log-likelihood.
![]() to the newly formed
covariance estimate. But, because we do not work
with
to the newly formed
covariance estimate. But, because we do not work
with
![]() directly, it is necessary
to implement the conditioning as follows: Let
directly, it is necessary
to implement the conditioning as follows: Let
![]() .
The upper triangular matrix
.
The upper triangular matrix ![]() is retained
and
is retained
and ![]() is discarded. Next,
we form the matrix as shown below for the case
is discarded. Next,
we form the matrix as shown below for the case ![]() :
:
![$\displaystyle {\bf R}^* =
\left[ \begin{array}{c} {\bf R} \cdots {\rm dia...
...
\rho_1 & 0 & 0 \\
0 & \rho_2 & 0 \\
0 & 0 & \rho_3
\end{array} \right]
$](img1453.png)
![]() where
where
![]() is an arbitrary covariance.
Let the eigendecomposition
of
is an arbitrary covariance.
Let the eigendecomposition
of
![]() be
be
![]() .
Clearly, then
.
Clearly, then
![\includegraphics[scale=0.66, clip]{cb.eps}](img1464.png)
![\includegraphics[width=3.2in,height=3.2in, clip]{br.eps}](img1465.png)
![\includegraphics[width=3.2in,height=3.2in, clip]{cr.eps}](img1466.png)